The Ultra II DNA Library Prep Kit for Illumina meets the challenge of constructing high quality libraries from ever-decreasing input quantities. The reagents for each step in the library preparation workflow have been reformulated to enable high yield preparation of high quality libraries from 500 picograms to 1 microgram of input DNA. This new generation of NEBNext reagents uses a fast, streamlined, automatable workflow and enables use of fewer PCR cycles while also improving GC coverage. The kit is also compatible with PCR-free workflows and is effective with challenging samples such as FFPE DNA.
Now also available with optional SPRIselect® beads
for clean-up and size-selection steps.
Advantages
-
Get more of what you need, with the highest library yields
-
Use to generate high quality libraries even when you have only limited amounts of DNA, with inputs as low as 500 pg
-
Prepare libraries from ALL of your samples, including GC-rich targets and FFPE DNA samples
-
Improve yields and quality for target enrichment applications
-
Save time with streamlined workflows , reduced hands-on time and automation compatibility, and enjoy the flexibility of kit or module formats
To learn more about how NEBNext Ultra II addresses lower input amounts and challenging sample types, download our technical note .
For use with NEBNext Multiplex Oligos for Illumina (Unique Dual Index UMI Adaptors DNA Set 1) (NEB #E7395 ), refer to the Protocols tab for UMI Adaptors-specific guidance.
NEBNext Ultra II produces the highest library yields and conversion rates from a broad range of input amounts
 A:
A: Libraries were prepared from Human NA19240 genomic DNA using the input amounts and numbers of PCR cycles shown. Manufacturers’ recommendations were followed, with the exception that size selection was omitted.
B: Libraries were prepared from Human NA19240 genomic DNA using the input amounts and library prep kits shown without an amplification step, following manufacturers' recommendations. qPCR was used to quantitate adaptor-ligated molecules, and quantitation values were then normalized to the conversion rate for Ultra II. The Ultra II Kit produces the highest rate of conversion to adaptor-ligated molecules, for a broad range of input amounts.
View additional data on library yield
NEBNext Ultra II provides uniform GC coverage for microbial genomic DNA over a broad range of GC composition and input amounts
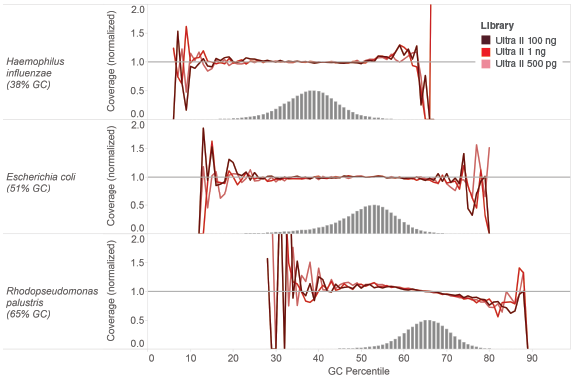
Libraries were made using 500 pg, 1 ng and 100 ng of the genomic DNAs shown and the Ultra II DNA Library Prep Kit and sequenced on an Illumina MiSeq
®. Reads were mapped using Bowtie 2.2.4 and GC coverage information was calculated using Picard's CollectGCBiasMetrics (v1.117). Expected normalized coverage of 1.0 is indicated by the horizontal grey line, the number of 100 bp regions at each GC% is indicated by the vertical grey bars, and the colored lines represent the normalized coverage for each library.
View additional data on library quality
NEBNext Ultra II provides superior yields in PCR-free workflows
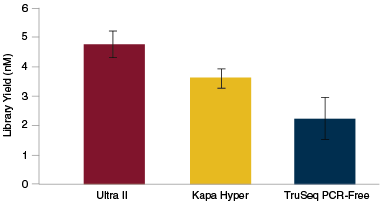
Libraries were generated from 100 ng of Human NA19240 genomic DNA using the library prep kits shown, following manufacturers' recommendations, and with no amplification step. Library yields were determined by qPCR using the NEBNext Library Quant Kit for Illumina. The NEBNext Ultra II Kit produces the highest yields.
NEBNext Ultra II libraries provide the highest quality sequence data in PCR-free workflows
|
LIBRARY KIT
|
TOTAL READS
|
% DUPLICATION
|
% MAPPED
|
% CHIMERAS
|
|
Ultra II
|
3,685,029
|
0.02
|
97.00
|
1.10
|
|
Kapa Hyper
|
3,679,136
|
0.02
|
97.00
|
2.00
|
|
TruSeq PCR-Free
|
3,681,643
|
0.05
|
96.65
|
1.38
|
PCR-free libraries were generated from 100 ng of Human NA19240 genomic DNA using library prep kits shown, following manufacturers’ recommendations, and with no amplification step. Libraries were sequenced on the Illumina NextSeq 500. Reads were mapped to the GRCh37 reference using Bowtie 2.2.4. This data illustrates that NEBNext Ultra II DNA Library Prep Kit enables high quality sequence data in PCR-free workflows, even with low input amounts.
% Mapped: The percentage of reads mapped to human GRCh37 reference.
% Duplication: The percentage of mapped sequence that is marked as duplicate.
% Chimeras: The percentage of reads that map outside of a maximum insert size or that have the two ends mapping to different chromosomes.
NEBNext Ultra II enables construction of high quality libraries from FFPE DNA samples
|
FFPE DNA
|
DNA INPUT (ng)
|
LIBRARY YIELDS IN ng
|
% MAPPED
|
% MAPPED IN PAIRS
|
% DUPLICATION
|
% CHIMERAS
|
|
Kidney Tumor
|
17
|
132
|
91.5
|
96.1
|
0.48
|
3.0
|
|
Lung Tumor
|
20
|
232
|
90.1
|
94.9
|
0.42
|
4.1
|
|
Liver Normal
|
20
|
691
|
92.6
|
94.7
|
0.33
|
8.6
|
|
Breast Tumor
|
30
|
514
|
91.9
|
95.1
|
0.37
|
4.5
|
Libraries were prepared from 17–30 ng of human DNA extracted from the FFPE tissue samples listed, amplified using 10 cycles of PCR and sequenced on the Illumina MiSeq.
Reads were mapped to the GRCh37 reference genome using Bowtie 2.2.4.
% Mapped: The percentage of reads mapped to Human GRCh37 reference.
% Mapped in Pairs: The percentage of reads whose mate pair was also aligned to the reference.
% Duplication: The percentage of mapped sequence that is marked as duplicate.
% Chimeras: The percentage of reads that map outside of a maximum insert size or that have the two ends mapping to different chromosomes.
This data illustrates that the NEBNext Ultra II DNA Library Prep Kit for Illumina enables high quality sequence data, even with low input amounts of FFPE DNA.
View additional data from FFPE DNA samples
NEBNext Ultra II provides superior yields for target enrichment applications.
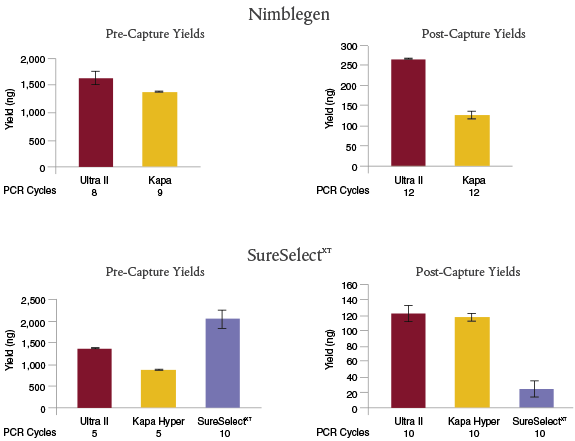
For Nimblegen
® enrichment, libraries were prepared with 100 ng of Human HG02922 genomic DNA, using NEBNext Ultra II with the NEBNext Adaptor and index primers, or the Kapa Library Preparation Kit Illumina Platforms with the SeqCap
® Adapter Kit (Roche). Target enrichment was performed with NimbleGen SeqCap Human Exome v3.
For SureSelect
XT enrichment, libraries were prepared with 200 ng of Human HG02922 genomic DNA, using NEBNext Ultra II or Kapa Hyper with the NEBNext Adaptor and index primers, or the SureSelect
XT Reagent Kit, Illumina (ILM) platforms with Herculase
® II Fusion DNA Polymerase. Target enrichment was performed with SureSelect
® Human All Exon V6.
Manufacturers’ recommendations were followed, including use of the recommended number of PCR cycles for pre- and post-capture. NEBNext Ultra II libraries resulted in the highest post-capture yields for both enrichment workflows.


 A: Libraries were prepared from Human NA19240 genomic DNA using the input amounts and numbers of PCR cycles shown. Manufacturers’ recommendations were followed, with the exception that size selection was omitted.
A: Libraries were prepared from Human NA19240 genomic DNA using the input amounts and numbers of PCR cycles shown. Manufacturers’ recommendations were followed, with the exception that size selection was omitted. Libraries were made using 500 pg, 1 ng and 100 ng of the genomic DNAs shown and the Ultra II DNA Library Prep Kit and sequenced on an Illumina MiSeq®. Reads were mapped using Bowtie 2.2.4 and GC coverage information was calculated using Picard's CollectGCBiasMetrics (v1.117). Expected normalized coverage of 1.0 is indicated by the horizontal grey line, the number of 100 bp regions at each GC% is indicated by the vertical grey bars, and the colored lines represent the normalized coverage for each library.
Libraries were made using 500 pg, 1 ng and 100 ng of the genomic DNAs shown and the Ultra II DNA Library Prep Kit and sequenced on an Illumina MiSeq®. Reads were mapped using Bowtie 2.2.4 and GC coverage information was calculated using Picard's CollectGCBiasMetrics (v1.117). Expected normalized coverage of 1.0 is indicated by the horizontal grey line, the number of 100 bp regions at each GC% is indicated by the vertical grey bars, and the colored lines represent the normalized coverage for each library.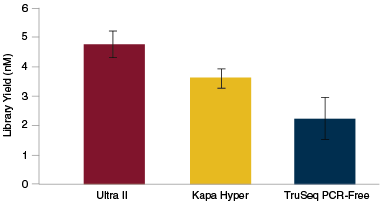 Libraries were generated from 100 ng of Human NA19240 genomic DNA using the library prep kits shown, following manufacturers' recommendations, and with no amplification step. Library yields were determined by qPCR using the NEBNext Library Quant Kit for Illumina. The NEBNext Ultra II Kit produces the highest yields.
Libraries were generated from 100 ng of Human NA19240 genomic DNA using the library prep kits shown, following manufacturers' recommendations, and with no amplification step. Library yields were determined by qPCR using the NEBNext Library Quant Kit for Illumina. The NEBNext Ultra II Kit produces the highest yields.
